Note: This blogpost is replicated from the AWS Big Data Blog and can be found here.
As the cost of genomic sequencing has rapidly decreased, the amount of publicly available genomic data has soared over the past couple of years. New cohorts and studies have produced massive datasets consisting of over 100,000 individuals. Simultaneously, these datasets have been processed to extract genetic variation across populations, producing mass amounts of variation data for each cohort.
In this era of big data, tools like Apache Spark have provided a user-friendly platform for batch processing of large datasets. However, to use such tools as a sufficient replacement to current bioinformatics pipelines, we need more accessible and comprehensive APIs for processing genomic data. We also need support for interactive exploration of these processed datasets.
ADAM and Mango provide a unified environment for processing, filtering, and visualizing large genomic datasets on Apache Spark. ADAM allows you to programmatically load, process, and select raw genomic and variation data using Spark SQL, an SQL interface for aggregating and selecting data in Apache Spark. Mango supports the visualization of both raw and aggregated genomic data in a Jupyter notebook environment, allowing you to draw conclusions from large datasets at multiple resolutions.
With the combined power of ADAM and Mango, you can load, query, and explore datasets in a unified environment. You can interactively explore genomic data at a scale previously impossible using single node bioinformatics tools. In this post, we describe how to set up and run ADAM and Mango on Amazon EMR. We demonstrate how you can use these tools in an interactive notebook environment to explore the 1000 Genomes dataset, which is publicly available in Amazon S3 as a public dataset.
Configuring ADAM and Mango on Amazon EMR
First, you launch and configure an EMR cluster. Mango uses Docker containers to easily run on Amazon EMR. Upon cluster startup, EMR uses the following bootstrap action to install Docker and the required startup scripts. The scripts are available at /home/hadoop/mango-scripts.
To start the Mango notebook, run the following:
This file sets up all of the environment variables that are needed to run Mango in Docker on Amazon EMR. In your terminal, you will see the port and Jupyter notebook token for the Mango notebook session. Navigate to this port on the public DNS URL of the master node for your EMR cluster.
Loading data from the 1000 Genomes Project
Now that you have a working environment, you can use ADAM and Mango to discover interesting variants in the child from the genome sequencing data of a trio (data from a mother, father, and child). This data is available from the 1000 Genomes Project AWS public dataset. In this analysis, you will view a trio (NA19685, NA19661, and NA19660) and search for variants that are present in the child but not present in the parents.
In particular, we want to identify genetic variants that are found in the child but not in the parents, known as de novo variants. These are interesting regions, as they can indicate sights of de novo variation that might contribute to multiple disorders.
You can find the Jupyter notebook containing these examples in Mango’s GitHub repository, or at /opt/cgl-docker-lib/mango/example-files/notebooks/aws-1000genomes.ipynb in the running Docker container for Mango.
First, import the ADAM and Mango modules and any Spark modules that you need:
Next, create a Spark session. You will use this session to run SQL queries on variants.
Variant analysis with Spark SQL
Load in a subset of variant data from chromosome 17:
You can take a look at the schema by printing the columns in the dataframe.
This genotypes dataset contains all samples from the 1000 Genomes Project. Therefore, you will next filter genotypes to only consider samples that are in the NA19685 trio, and cache the results in memory.
Next, add a new column to your dataframe that determines the genomic location of each variant. This is defined by the chromosome (contigName) and the start and end position of the variant.
Now, you can query your dataset to find de novo variants. But first, you must register your dataframe with Spark SQL.
Now that your dataframe is registered, you can run SQL queries on it. For the first query, select the names of variants belonging to sample NA19685 that have at least one alternative (ALT) allele.
For your next query, filter sites in which the parents have both reference alleles. Then filter these variants by the set produced previously from the child.
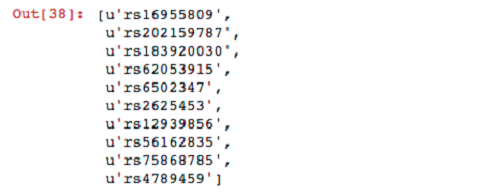
Now that you have found some interesting variants, you can unpersist your genotypes from memory.
Working with alignment data
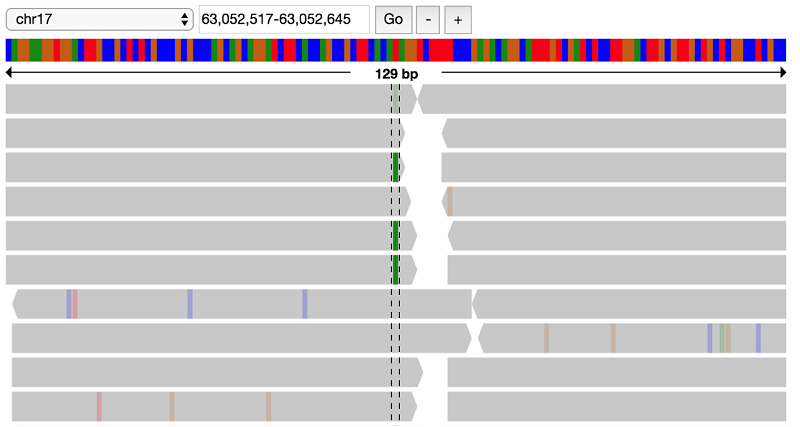
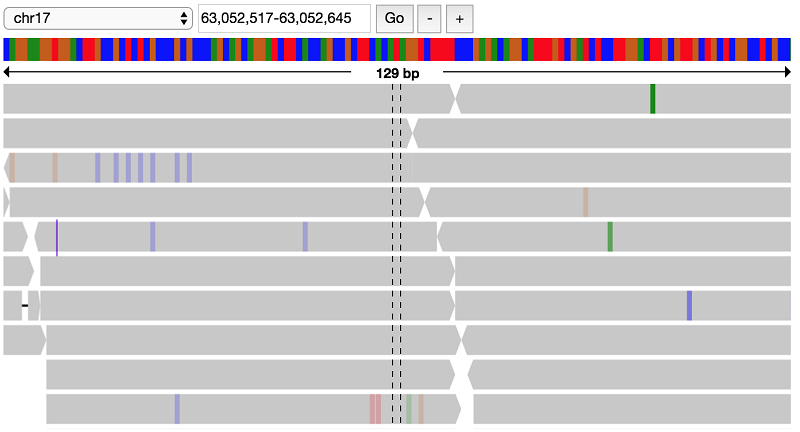
You have found a lot of potential de novo variant sites. Next, you can visually verify some of these sites to see if the raw alignments match up with these de novo hits.
First, load in the alignment data for the NA19685 trio:
Note that this example uses s3a:// instead of s3:// style URLs. The reason for this is that the ADAM formats use Java NIO to access BAM files. To do this, we are using a JSR 203implementation for the Hadoop Distributed File System to access these files. This itself requires the s3a:// protocol. You can view that implementation in this GitHub repository.
You now have data alignment data for three individuals in your trio. However, the data has not yet been loaded into memory. To cache these datasets for fast subsequent access to the data, run the cache() function:
Quality control of alignment data
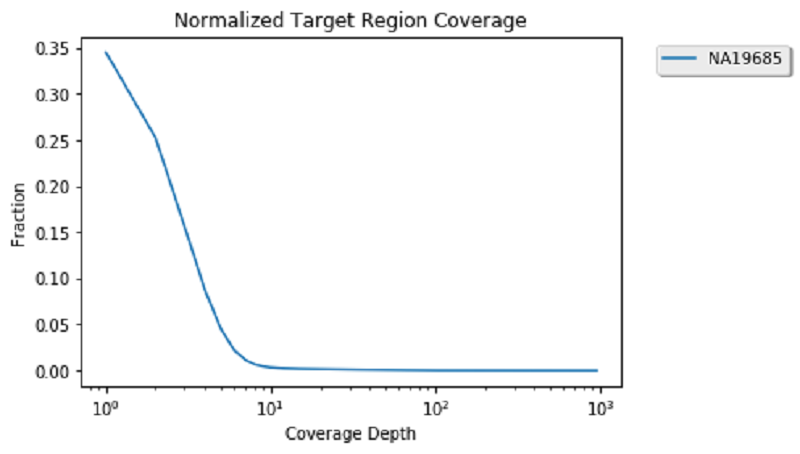
One popular analysis to visually re-affirm the quality of genomic alignment data is by viewing coverage distribution. Coverage distribution gives you an idea of the read coverage that you have across a sample.
Next, generate a sample coverage distribution plot for the child alignment data on chromosome 17: